This gem removes common margin from indented strings, such as the ones produced by indented heredocs. In other words, it strips out leading whitespace chars at the beginning of each line, but only as much as the line with the smallest margin.
It is acknowledged that many strings defined by heredocs are just code and fact is that most parsers are insensitive to indentation. If, however, the strings are to be used otherwise, be it for printing or testing, the extra indentation will probably be an issue and hence this gem.
This is a pedagogical package, designed to help students understanding convergence of random variables. It provides a way to investigate interactively various modes of convergence (in probability, almost surely, in law and in mean) of a sequence of i.i.d. random variables. Visualisation of simulated sample paths is possible through interactive plots. The approach is illustrated by examples and exercises through the function investigate', as described in Lafaye de Micheaux and Liquet (2009) <doi:10.1198/tas.2009.0032>. The user can study his/her own sequences of random variables.
This package provides SPSS'- and SAS'-like output for cross tabulations of two categorical variables (CROSSTABS) and for hierarchical loglinear analyses of two or more categorical variables (LOGLINEAR). The methods are described in Agresti (2013, ISBN:978-0-470-46363-5), Ajzen & Walker (2021, ISBN:9780429330308), Field (2018, ISBN:9781526440273), Norusis (2012, ISBN:978-0-321-74843-0), Nussbaum (2015, ISBN:978-1-84872-603-1), Stevens (2009, ISBN:978-0-8058-5903-4), Tabachnik & Fidell (2019, ISBN:9780134790541), and von Eye & Mun (2013, ISBN:978-1-118-14640-8).
Engineered features and "helper" functions ancillary to the public.ctn0094data package, extending this package for ease of use (see <https://CRAN.R-project.org/package=public.ctn0094data>). This public.ctn0094data package contains harmonized datasets from some of the National Institute of Drug Abuse's Clinical Trials Network (NIDA's CTN) projects. Specifically, the CTN-0094 project is to harmonize and de-identify clinical trials data from the CTN-0027, CTN-0030, and CTN-51 studies for opioid use disorder. This current version is built from public.ctn0094data v. 1.0.6.
Scaling models and classifiers for sparse matrix objects representing textual data in the form of a document-feature matrix. Includes original implementations of Laver', Benoit', and Garry's (2003) <doi:10.1017/S0003055403000698>, Wordscores model, the Perry and Benoit (2017) <doi:10.48550/arXiv.1710.08963> class affinity scaling model, and the Slapin and Proksch (2008) <doi:10.1111/j.1540-5907.2008.00338.x> wordfish model, as well as methods for correspondence analysis, latent semantic analysis, and fast Naive Bayes and linear SVMs specially designed for sparse textual data.
This package provides methods for analysis of energy consumption data (electricity, gas, water) at different data measurement intervals. The package provides feature extraction methods and algorithms to prepare data for data mining and machine learning applications. Deatiled descriptions of the methods and their application can be found in Hopf (2019, ISBN:978-3-86309-669-4) "Predictive Analytics for Energy Efficiency and Energy Retailing" <doi:10.20378/irbo-54833> and Hopf et al. (2016) <doi:10.1007/s12525-018-0290-9> "Enhancing energy efficiency in the residential sector with smart meter data analytics".
Many recursive functions share the same structure, e.g. pattern-match on the input and, depending on the data constructor, either recur on a smaller input or terminate the recursion with the base case. Another one: start with a seed value, use it to produce the first element of an infinite list, and recur on a modified seed in order to produce the rest of the list. Such a structure is called a recursion scheme. Using higher-order functions to implement those recursion schemes makes your code clearer, faster, and safer. See README for details.
Fits a piecewise exponential hazard to survival data using a Hierarchical Bayesian model with an Intrinsic Conditional Autoregressive formulation for the spatial dependency in the hazard rates for each piece. This function uses Metropolis- Hastings-Green MCMC to allow the number of split points to vary and also uses Stochastic Search Variable Selection to determine what covariates drive the risk of the event. This function outputs trace plots depicting the number of split points in the hazard and the number of variables included in the hazard. The function saves all posterior quantities to the desired path.
Data on standard load profiles from the German Association of Energy and Water Industries (BDEW Bundesverband der Energie- und Wasserwirtschaft e.V.) in a tidy format. The data and methodology are described in VDEW (1999), "Repräsentative VDEW-Lastprofile", <https://www.bdew.de/media/documents/1999_Repraesentative-VDEW-Lastprofile.pdf>. The package also offers an interface for generating a standard load profile over a user-defined period. For the algorithm, see VDEW (2000), "Anwendung der Repräsentativen VDEW-Lastprofile step-by-step", <https://www.bdew.de/media/documents/2000131_Anwendung-repraesentativen_Lastprofile-Step-by-step.pdf>.
This package provides a fully parameterized Generalized Wendland covariance function for use in Gaussian process models, as well as multiple methods for approximating it via covariance interpolation. The available methods are linear interpolation, polynomial interpolation, and cubic spline interpolation. Moreno Bevilacqua and Reinhard Furrer and Tarik Faouzi and Emilio Porcu (2019) <url:<https://projecteuclid.org/journalArticle/Download?urlId=10.1214%2F17-AOS1652 >>. Moreno Bevilacqua and Christian Caamaño-Carrillo and Emilio Porcu (2022) <doi:10.48550/arXiv.2008.02904>. Reinhard Furrer and Roman Flury and Florian Gerber (2022) <url:<https://CRAN.R-project.org/package=spam >>.
The classical Markowitz's mean-variance portfolio formulation ignores heavy tails and skewness. High-order portfolios use higher order moments to better characterize the return distribution. Different formulations and fast algorithms are proposed for high-order portfolios based on the mean, variance, skewness, and kurtosis. The package is based on the papers: R. Zhou and D. P. Palomar (2021). "Solving High-Order Portfolios via Successive Convex Approximation Algorithms." <arXiv:2008.00863>. X. Wang, R. Zhou, J. Ying, and D. P. Palomar (2022). "Efficient and Scalable High-Order Portfolios Design via Parametric Skew-t Distribution." <arXiv:2206.02412>.
Calculate the injury severity score (ISS) based on the dictionary in ICDPIC from <https://ideas.repec.org/c/boc/bocode/s457028.html>. The original code was written in STATA 11'. The original STATA code was written by David Clark, Turner Osler and David Hahn. I implement the same logic for easier access. Ref: David E. Clark & Turner M. Osler & David R. Hahn, 2009. "ICDPIC: Stata module to provide methods for translating International Classification of Diseases (Ninth Revision) diagnosis codes into standard injury categories and/or scores," Statistical Software Components S457028, Boston College Department of Economics, revised 29 Oct 2010.
The ultimate goal is to support 2-2-1, 2-1-1, and 1-1-1 models for multilevel mediation, the option of a moderating variable for either the a, b, or both paths, and covariates. Currently the 1-1-1 model is supported and several options of random effects; the initial code for bootstrapping was evaluated in simulations by Falk, Vogel, Hammami, and MioÄ eviÄ (2024) <doi:10.3758/s13428-023-02079-4>. Support for Bayesian estimation using brms comprises ongoing work. Currently only continuous mediators and outcomes are supported. Factors for any predictors must be numerically represented.
The ClusterSignificance package provides tools to assess if class clusters in dimensionality reduced data representations have a separation different from permuted data. The term class clusters here refers to, clusters of points representing known classes in the data. This is particularly useful to determine if a subset of the variables, e.g. genes in a specific pathway, alone can separate samples into these established classes. ClusterSignificance accomplishes this by, projecting all points onto a one dimensional line. Cluster separations are then scored and the probability of the seen separation being due to chance is evaluated using a permutation method.
Rapid is a Go library for property-based testing.
Rapid checks that properties you define hold for a large number of automatically generated test cases. If a failure is found, rapid automatically minimizes the failing test case before presenting it.
Features:
imperative Go API with type-safe data generation using generics
data generation biased to explore "small" values and edge cases more thoroughly
fully automatic minimization of failing test cases
persistence and automatic re-running of minimized failing test cases
support for state machine ("stateful" or "model-based") testing
no dependencies outside the Go standard library
This plugin goes to great lengths to save and restore all the details from your tmux environment. Here's what's been taken care of:
all sessions, windows, panes and their order
current working directory for each pane
exact pane layouts within windows (even when zoomed)
active and alternative session
active and alternative window for each session
windows with focus
active pane for each window
"grouped sessions" (useful feature when using tmux with multiple monitors)
programs running within a pane! More details in the restoring programs doc.
Optional:
restoring vim and neovim sessions
restoring pane contents
Computes the key metrics for assessing the performance of a liquidity provider (LP) position in a weighted multi-asset Automated Market Maker (AMM) pool. Calculates the nominal and percentage impermanent loss (IL) by comparing the portfolio value inside the pool (based on the weighted geometric mean of price ratios) against the value of simply holding the assets outside the pool (based on the weighted arithmetic mean). The primary function, `impermanent_loss()`, incorporates the effect of earned trading fees to provide the LP's net profit and loss relative to a holding strategy, using a methodology derived from Tiruviluamala, N., Port, A., and Lewis, E. (2022) <doi:10.48550/arXiv.2203.11352>.
This package provides several measures ((dis)similarity, distance/metric, correlation, entropy) for comparing two partitions of the same set of objects. The different measures can be assigned to three different classes: Pair comparison (containing the famous Jaccard and Rand indices), set based, and information theory based. Many of the implemented measures can be found in Albatineh AN, Niewiadomska-Bugaj M and Mihalko D (2006) <doi:10.1007/s00357-006-0017-z> and Meila M (2007) <doi:10.1016/j.jmva.2006.11.013>. Partitions are represented by vectors of class labels which allow a straightforward integration with existing clustering algorithms (e.g. kmeans()). The package is mostly based on the S4 object system.
Two arms clinical trials required sample size is calculated in the comprehensive parametric context. The calculation is based on the type of endpoints(continuous/binary/time-to-event/ordinal), design (parallel/crossover), hypothesis tests (equality/noninferiority/superiority/equivalence), trial arms noncompliance rates and expected loss of follow-up. Methods are described in: Chow SC, Shao J, Wang H, Lokhnygina Y (2017) <doi:10.1201/9781315183084>, Wittes, J (2002) <doi:10.1093/epirev/24.1.39>, Sato, T (2000) <doi:10.1002/1097-0258(20001015)19:19%3C2689::aid-sim555%3E3.0.co;2-0>, Lachin J M, Foulkes, M A (1986) <doi:10.2307/2531201>, Whitehead J(1993) <doi:10.1002/sim.4780122404>, Julious SA (2023) <doi:10.1201/9780429503658>.
Calculates sample size for various scenarios, such as sample size to estimate population proportion with stated absolute or relative precision, testing a single proportion with a reference value, to estimate the population mean with stated absolute or relative precision, testing single mean with a reference value and sample size for comparing two unpaired or independent means, comparing two paired means, the sample size For case control studies, estimating the odds ratio with stated precision, testing the odds ratio with a reference value, estimating relative risk with stated precision, testing relative risk with a reference value, testing a correlation coefficient with a specified value, etc. <https://www.academia.edu/39511442/Adequacy_of_Sample_Size_in_Health_Studies#:~:text=Determining%20the%20sample%20size%20for,may%20yield%20statistically%20inconclusive%20results.>.
Computation of sparse portfolios for financial index tracking, i.e., joint selection of a subset of the assets that compose the index and computation of their relative weights (capital allocation). The level of sparsity of the portfolios, i.e., the number of selected assets, is controlled through a regularization parameter. Different tracking measures are available, namely, the empirical tracking error (ETE), downside risk (DR), Huber empirical tracking error (HETE), and Huber downside risk (HDR). See vignette for a detailed documentation and comparison, with several illustrative examples. The package is based on the paper: K. Benidis, Y. Feng, and D. P. Palomar, "Sparse Portfolios for High-Dimensional Financial Index Tracking," IEEE Trans. on Signal Processing, vol. 66, no. 1, pp. 155-170, Jan. 2018. <doi:10.1109/TSP.2017.2762286>.
This package provides a simple interface to the Geographic Header information from the "2010 US Census Summary File 2". The entire Summary File 2 is described at <https://catalog.data.gov/dataset/census-2000-summary-file-2-sf2>, but note that this package only provides access to parts of the geographic header ('geoheader') of the file. In particular, only the first 101 columns of the geoheader are included and, more importantly, only rows with summary levels (SUMLEVs) 010 through 050 (nation down through county level) are included. In addition to access to (part of) the geoheader, the package also provides a decode function that takes a column name and value and, for certain columns, returns "the meaning" of that column (i.e., a "SUMLEV" value of 40 means "State"); without a value, the decode function attempts to describe the column itself.
An exact method for computing the Poisson-Binomial Distribution (PBD). The package provides a function for generating a random sample from the PBD, as well as two distinct approaches for computing the density, distribution, and quantile functions of the PBD. The first method uses direct-convolution, or a dynamic-programming approach which is numerically stable but can be slow for a large input due to its quadratic complexity. The second method is much faster on large inputs thanks to its use of Fast Fourier Transform (FFT) based convolutions. Notably in this case the package uses an exponential shift to practically guarantee the relative accuracy of the computation of an arbitrarily small tail of the PBD -- something that FFT-based methods often struggle with. This ShiftConvolvePoiBin method is described in Peres, Lee and Keich (2020) <arXiv:2004.07429> where it is also shown to be competitive with the fastest implementations for exactly computing the entire Poisson-Binomial distribution.
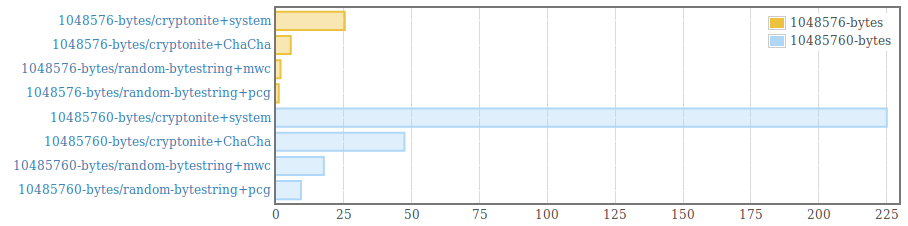
__This package is deprecated__. Please, use genByteString from the [random package (version >=1.2)](https://hackage.haskell.org/package/random) instead. . Efficient generation of random bytestrings. The implementation populates uninitialized memory with uniformily distributed random 64 bit words (and 8 bit words for remaining bytes at the end of the bytestring). . Random words are generated using the PRNG from the [mwc-random](https://hackage.haskell.org/package/mwc-random) package or the [pcg-random](https://hackage.haskell.org/package/pcg-random) package. It is also possible to use a custom PRNG by providing an instance for the RandomWords type class and using the function generate from the module "Data.ByteString.Random.Internal". . The generated byte strings are suitable for statistical applications. They are /not/ suitable for cryptographic applications. . 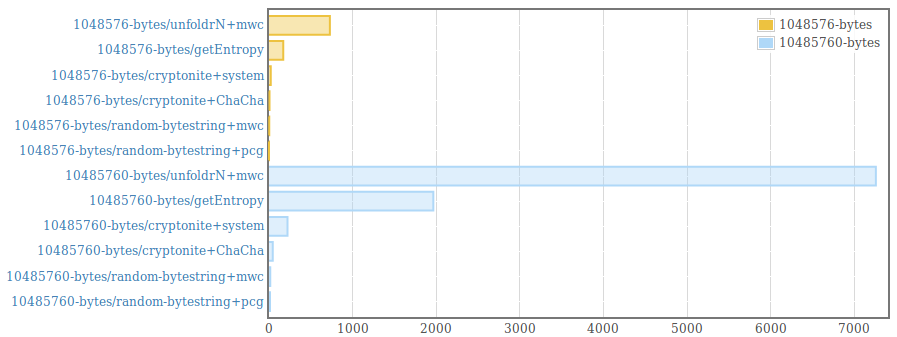 .